There was a really important scientific result reported on this week![]() in the press. The original paper
in the press. The original paper![]() , by a team at Scripps Research Institute in La Jolla, CA, a person in Grenoble, France, and a person in Henan, China, is behind a paywall at the National Academy of Science.
, by a team at Scripps Research Institute in La Jolla, CA, a person in Grenoble, France, and a person in Henan, China, is behind a paywall at the National Academy of Science.
This team had previously introduced a new, unnatural base pair (UPB) into the DNA of an organism based on E. coli. In the past it had caused some toxicity to the organism and also tended to get deleted during reproduction. The new result is that they synthetically modified the organism, getting rid of the toxicity, and showed that the UBP could survive 60 generations of reproduction.
Here is what normal DNA (deoxyribonucleic acid) looks like (from Wikimedia Commons):
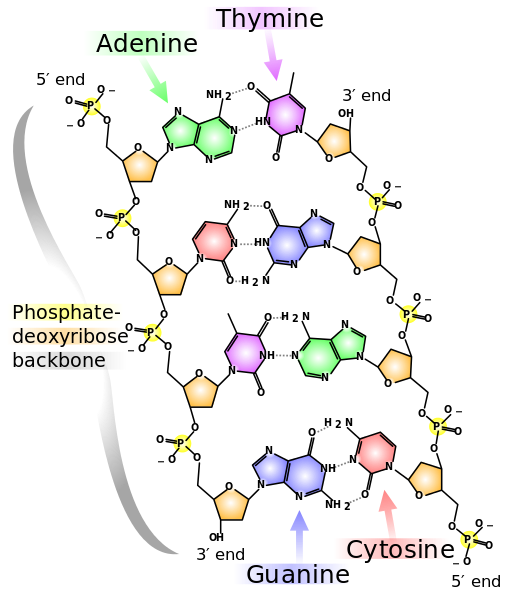
There are two backbone chains, left and right, of alternating 2-deoxyribose and phosphate molecules joined by complementary pairs of nucleotide pairs of either Adeline (A) and Thymine (T) or of Guanine (G) and Cytosine (C). So reading down the left side of this fragment of DNA we have the code ACTG, and reading up the right side we have CAGT.
There are lots of mechanisms about DNA and RNA that are not fully understood still, but DNA is used for two purposes. The letters on it encode genetic sequences which are used to construct proteins (it gets more complex every decade as we understand more), the stuff of life, and it is used to make copies of itself so that one copy can remain in a parent cell and another copy goes to a new child cells.
For producing proteins the two strands or backbones are pried apart with a molecular machine moving along it, and and RNA molecule is built with complementary base pairs for sub-length of the DNA. RNA (ribonucleic acid) looks like this, with just one backbone chain where ribose (which has five Oxygen atoms rather than the four of deoxyribose) molecules and phosphate molecules alternate and single bases, one of the four letters, hang off at regular intervals.
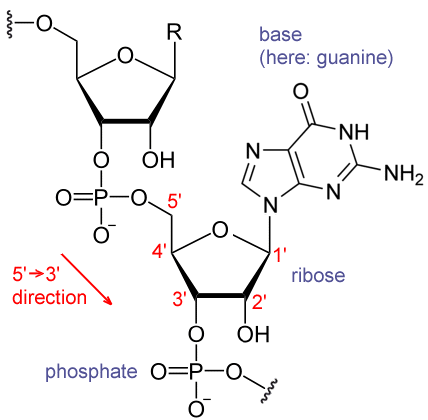
The process of producing this RNA in this way is know as transcription. It then gets translated by another mechanism into amino acids which are linked together to produce proteins. In all life on earth the series of letters is used three at a time (which means 64 possible combinations of the four letters ![]() ) of which in the “standard” setting 61 of the codings select for one of 20 amino acids, and the remaining three codings are used to say stop. These 64 cases can easily be written down as a table for all the possible three letter sequences (which themselves are known as codons). There are currently close to 30 (numbers change all the time…) variations on this code found in life on Earth–for instance vertebrates, invertebrates, and yeasts, each use their own slightly different version of the table in translating the DNA in the mitochondria of their cells, coding for a total of 23 amino acids (I think…)
) of which in the “standard” setting 61 of the codings select for one of 20 amino acids, and the remaining three codings are used to say stop. These 64 cases can easily be written down as a table for all the possible three letter sequences (which themselves are known as codons). There are currently close to 30 (numbers change all the time…) variations on this code found in life on Earth–for instance vertebrates, invertebrates, and yeasts, each use their own slightly different version of the table in translating the DNA in the mitochondria of their cells, coding for a total of 23 amino acids (I think…)
But here is a thing one; since 1990 people have done experiments where they have modified simple organisms to change the meanings of some codons to produce amino acids (there are many of them known in nature) which are not coded for in any natural system. We will come back to this.
The second thing that happens to DNA is in reproduction and that works as follows. The double stranded DNA is fed into a little molecular machine which unzips it where the base pairs join, and then lets a complementary base and newly constructed backbone attach to each half of the DNA, spitting out, in a continuous fashion two copies of the original DNA, where each copy has half of the actual atoms of the original.
Now what does this new paper do? It has added a new pair of bases to an E. coli genome, and built a version of E. coli where that reproduction mechanism for DNA handles the new letters well, and where they existence of the new letters causes no real harm to the cell.
We can call the new bases by the letters X and Y, though as you can see from this diagram they have longer names. This is figure 1A from the paper:
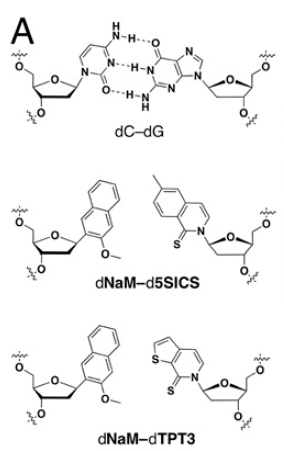
At the top we see a standard Cytosine-Guanine pair, and below that two variations of X and Y (the same X in the two cases) pairings. In this later paper they have shown that they can build a robust semi synthetic organism that carries these X and Y letters in the DNA, and preserve those letters well over at least 60 generations–that means at least 60 consecutive zippings apart and copying of the DNA including the X’s and Y’s. In one variation they experiment with all 16 possible three letter sequences which have X in the middle and one of the regular G, A, C, or T on either side. They state that the “loss was minimal to undetectable in 13 of the 16 cases”.
For my commentary below lets call this thing two. We have now seen unnatural base pairs in a living organism being reproduced reliably.
Now the next thing that one imagines these scientists must be excited about is getting the transcription mechanism to handle the new letters, and then expanding the translation table from ![]() entries to some bigger number. The theoretical maximum would be
entries to some bigger number. The theoretical maximum would be ![]() , though so far they have not shown any sequences that have X’s or Y’s adjacent to each other are preserved. But let’s call this combined result of two mechanisms thing three.
, though so far they have not shown any sequences that have X’s or Y’s adjacent to each other are preserved. But let’s call this combined result of two mechanisms thing three.
Thing one and thing two have been demonstrated. Thing three has not.
But why am I writing this post. It is because I think thing two is a big deal about what life elsewhere might look like.
There has been some debate over whether life everywhere might look at the molecular level just like life here on Earth. I.e., perhaps it is the case that there is only a one way to make life out the the chemistry that exists in our Universe (and we assume here for argument’s sake that chemistry is the same everywhere in the Universe though there is debate about that).
We already thought, due to the multiple natural translation tables in Earth life, admittedly small variations on each other, but also that thing one had been done and varied them further that it might be reasonable to expect life, if we ever find it, elsewhere in the Solar System of further afield, to have different translation tables. In fact that has been a key question if we were to find life on Mars. If it has the same translation tables as on Earth we might presume that both forms of life came from the same place, perhaps Mars. We have identified many meteorites on Earth that were once part of Mars, blasted off the surface of Mars by a large impact and eventually falling to Earth millions of years later. Perhaps they brought life with them. But if we found DNA-based life on Mars to have a very different translation table from that on Earth we would tend to think that the life had arisen twice independently.
Now with thing two having been demonstrated in this new paper we might expect DNA based life on Mars to be even more different than that on Earth, perhaps use]ing a different set of base pairs. Since we have XY and XY’ demonstrated in this paper, we could imagine that it is not such a big step to have life with none of GACT, but perhaps all based in XYZW, or PQRS, or perhaps IJKLMN. This opens up the possibilities mightily. It is no longer enough to assay samples from Mars for the four base nucleotides that we find on Earth and declare no life if we do not see them. Before we get ahead of ourselves however, we must wait for thing three to be demonstrated. But that will seal the fate of how we must look for life on Mars–in a much more expansive way.
Is there a thing four? Yes, perhaps in another version of DNA/RNA based biology there are not three letters used for each amino acid. In a simpler version there might be only two letters to determine a smaller number of possible amino acids, or in a more complex version four letters to determine a larger number. The engineering challenges to modify Earth based life to perform this way are significant, so I would not expect to see that any time soon. But it could have implications for life elsewhere.
Getting back to Earth biology people have been trying to understand how RNA and DNA showed up to make life anywhere. A fairly sure bet is that there were simpler mechanisms before the current mechanisms we see. Perhaps all that life got obliterated, competed away, by the much more stable RNA/DNA based life we see today. Or perhaps some of it is still hiding in isolated environments on Earth and we haven’t yet recognized it.
One hypothesis is that perhaps a much less stable form of life relied on the much simpler PNA (peptide nucleic acid) shown here, but using the same modern GACT.
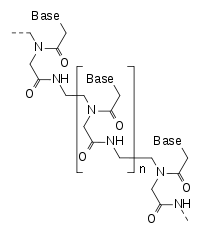
This is a much simpler backbone and there are arguments that it could more easily have arisen spontaneously in the primordial soup, but it is not as stable as DNA for long term storage of genetic information. People have been doing lab experiments for twenty years getting PNA with the standard GACT bases to interact with and transfer sequences with RNA and DNA. There are independently arguments about how the redundant standard translation table (61 coding entries but only 20 different amino acids), could have evolved from a much simpler coding system.
I think thing two shows that we must be more expansive on what we believe the biochemistry of life elsewhere might be.
My own suspicion is that there is plenty of life out there that uses totally different coding systems, and totally different molecules than RNA and DNA.
And I am getting more and more convinced that our current tools for detecting life are “all the harder to see you with”!
![]() This particular story has some questionable wording in places. This is not an entirely new type of DNA. Rather it is completely conventional DNA but it carries a new pair of base nucleotides.
This particular story has some questionable wording in places. This is not an entirely new type of DNA. Rather it is completely conventional DNA but it carries a new pair of base nucleotides.
![]() Yorke Zhang, Brian M. Lamb, Aaron W. Feldman, Anne Xiaozhou Zhou, Thomas Lavergne, Lingjun Li, and Floyd E. Romesberg, A semisynthetic organism engineered for the stable expansion of the genetic alphabet, Proceedings of the National Academy of Science, www.pnas.org/cgi/doi/10.1073/pnas.1616443114
Yorke Zhang, Brian M. Lamb, Aaron W. Feldman, Anne Xiaozhou Zhou, Thomas Lavergne, Lingjun Li, and Floyd E. Romesberg, A semisynthetic organism engineered for the stable expansion of the genetic alphabet, Proceedings of the National Academy of Science, www.pnas.org/cgi/doi/10.1073/pnas.1616443114

